
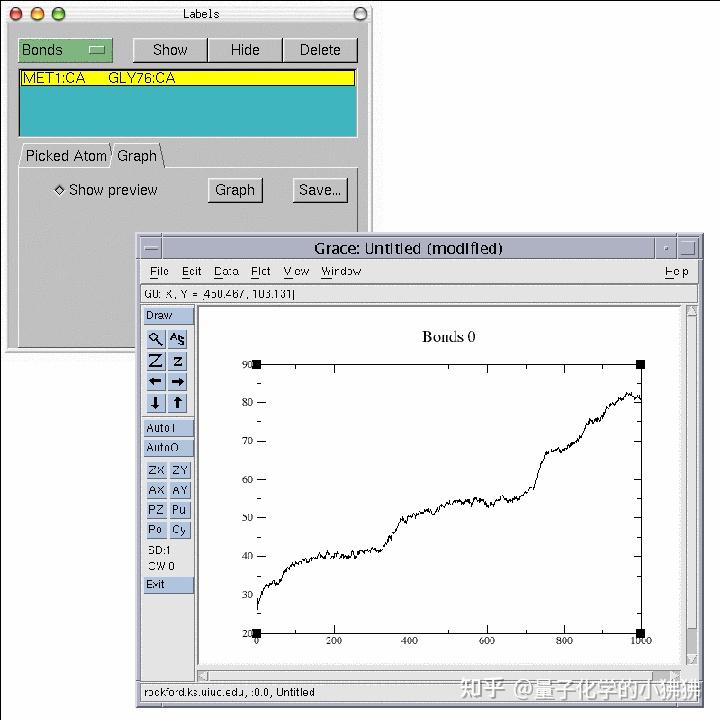
Menu, click on the button in the lower left.ĥ Choose Graphics Representations menu item. Will be explained extensively later in the tutorial. To go to theīeginning of the trajectory, you will use one of the Animation Tools, that You will be able to see the frames as they areĤ After the trajectory finishes loading, you will be will be looking at the last frame of your trajectory. Browse for pulling.dcd, click OK and click on the Load button again. The Browse button, making sure that ubiquitin.psf is The protein structure (PSF) file, so we need to load first the parameterįile, and then add the trajectory data to this file, as explained in Unit 2.Ģ Load the PSF file of the system ubiquitin.psf, as done inģ In the Molecule File Browser window, click on Trajectory files do not contain the information of the system contained in An example of trajectory files are DCD files. Trajectory files are normally binary files that contain several You will now learn to load the time evolving coordinates of a The ubiquitin system is equilibrated by looking at RMSD plots. Macros, place labels on atoms and bonds, and calculate the RMSD of a
VMD MEASURE RMSD HOW TO
In this unit, you will learn how to load trajectories, create An Example Tcl Script: Calculating the RMSD of a trajectory.Up: VMD Tutorial Previous: Multiple Molecules and Scripting


 0 kommentar(er)
0 kommentar(er)
